import pybliometrics
pybliometrics.scopus.utils.create_config()Learn to parse data from Scopus using Python.
Any sound scientific inquiry is grounded on a meticulous review of relevant literature. Today, we have multiple online repositories for storing and accessing literature from different disciplines. For instance, research articles in the domain of biological sciences can be accessed through databases like PubMed, Scopus, Web of Sciences, etc. The information available through these databases is of immense value to the research community. However, the exponential growth of these databases over the last few years has made it challenging to parse the new information. Also, modern research is demolishing the disciplinary divide such that research now needs to access and process literature from different sources. To address these issues we need computational methods to effectively search the online literature archives. Literature mining is an emerging field that enables researchers to programmatically parse text data (scientific literature) to not only extract the required information but also to generate testable hypotheses. Here, we’ll look into one of the Python libraries for mining literature from the Scopus database. The example provided here would act as a foundation for designing comprehensive and rigorous text-mining strategies.
Elsevier Developer Portal
The first thing you need to do is to get an API key to access the Elsevier resoucres programmatically (Link). The APIs are available for no-charge for non-commercial use. However, an important point to note is that if you are not a subscriber to Scopus then there’ll a limit of parsing upto 5,000 articles per search. On the other hand, if you’ve an active subscription then there is no such limit.
Pybliometrics library
Next, you need the pybliometrics library Link to access the Scopus database programmatically. The library can be install by running the command pip install pybliometrics. Once you have got the API keys and installed the pybliometrics library, the key must be saved in the configuration file (config.ini) as described here or you may execute the following two lines of code to create the configuration file.
We’ll now import AbstractRetrieval, AuthorRetrieval, and ScopusSearch functions from the pybliometrics.scopus class. These functions can be used to search Scopus based on doi, author id, or search string, respectively. We’ll also import pandas and matplotlib to facilitate parsing and visualization of the results. The AbstractRetrieval function takes a doi as an argument and return an object of the namesake class. This object has multiple attribute to get information about the document. E.g. the title attribute gives the title of the article.
from pybliometrics.scopus import AbstractRetrieval
from pybliometrics.scopus import AuthorRetrieval
from pybliometrics.scopus import ScopusSearch
import pandas as pd
import matplotlib.pyplot as pltab = AbstractRetrieval("10.1016/j.softx.2019.100263")
ab.title'pybliometrics: Scriptable bibliometrics using a Python interface to Scopus'Similarly, AuthorRetrieval function can be used to search Scopus using a Scopus author ID as an argument. The returned object has a get_documents function that can be used to generate a list having each publication as its element.
author_search = AuthorRetrieval("24398410800")
my_pubs = author_search.get_documents()ctr=0
while(ctr<3):
print(my_pubs[ctr].coverDate, my_pubs[ctr].title)
ctr+=12021-12-01 Interplay of substrate polymorphism and conformational plasticity of Plasmodium tyrosyl-tRNA synthetase
2021-07-15 Safranal inhibits NLRP3 inflammasome activation by preventing ASC oligomerization
2020-11-01 In silico assessment of natural products and approved drugs as potential inhibitory scaffolds targeting aminoacyl-tRNA synthetases from PlasmodiumCustomized search
To retrieve publications based on a customized search string, we can use the ScopusSearch function which takes as an argument a valid Scopus search query. For example, the code below shows a search string to search the Scopus database for all the publications having diabetes and Alzheimer’s in the title. The subscriber attribute for this function takes a Boolean value depending on the status of the subscription. The ScopusSearch function returns a ScopusSearch class object that has an attribute results that provides a list of all the documents matching the search criteria. In the example below, we’ll search the Scopus database using two different search strategies: - Search for all the publications having diabetes AND Alzheimer’s in the title of the article. - Search for all the publications having diabetes AND Parkinson’s in the title of the article.
search_dia_A = ScopusSearch("TITLE ( diabetes AND Alzheimer's ) ",subscriber=False)
search_dia_P = ScopusSearch("TITLE ( diabetes AND Parkinson's ) ",subscriber=False)type(search_dia_A)pybliometrics.scopus.scopus_search.ScopusSearchprint("Total number of articles for search_dia_A:", len(search_dia_A.results))
print("Total number of articles for search_dia_P:", len(search_dia_P.results))Total number of articles for search_dia_A: 689
Total number of articles for search_dia_P: 158Let’s make two dataframes have all the results from the two searches. The document object is a named tuple with information about the articles such as title, doi, authors, etc. Using this object we’ll create a dataframe for each of the documents and then concat all these dataframes. Note that while concatnating we’ll transpose the dataframe so that the keywords in the named tuple becomes the column headers for the final dataframe. We also need to explicitly name the columns of the concatenated dataframe.
search_dia_A.results[0]Document(eid='2-s2.0-85141971759', doi='10.1016/j.neuropharm.2022.109327', pii='S0028390822003860', pubmed_id='36368623', title="The common genes involved in the pathogenesis of Alzheimer's disease and type 2 diabetes and their implication for drug repositioning", subtype='ar', subtypeDescription='Article', creator='Yuan X.', afid=None, affilname='Tianjin Medical University', affiliation_city='Tianjin', affiliation_country='China', author_count=None, author_names=None, author_ids=None, author_afids=None, coverDate='2023-02-01', coverDisplayDate='1 February 2023', publicationName='Neuropharmacology', issn='00283908', source_id='20435', eIssn='18737064', aggregationType='Journal', volume='223', issueIdentifier=None, article_number='109327', pageRange=None, description=None, authkeywords=None, citedby_count=0, openaccess=0, freetoread=None, freetoreadLabel=None, fund_acr=None, fund_no=None, fund_sponsor=None)df_results_dia_A = pd.DataFrame()
for document in search_dia_A.results:
df_results_dia_A = pd.concat([df_results_dia_A, pd.DataFrame(document).T], axis=0)
df_results_dia_P = pd.DataFrame()
for document in search_dia_P.results:
df_results_dia_P = pd.concat([df_results_dia_P, pd.DataFrame(document).T], axis=0)df_results_dia_A.columns = ['eid', 'doi', 'pii', 'pubmed_id', 'title', 'subtype', 'subtypeDescription', 'creator', 'afid', 'affilname', 'affiliation_city', 'affiliation_country', 'author_count', 'author_names', 'author_ids', 'author_afids', 'coverDate', 'coverDisplayDate', 'publicationName', 'issn', 'source_id', 'eIssn', 'aggregationType', 'volume', 'issueIdentifier', 'article_number', 'pageRange', 'description', 'authkeywords', 'citedby_count', 'openaccess', 'freetoread', 'freetoreadLabel', 'fund_acr', 'fund_no', 'fund_sponsor']
df_results_dia_P.columns = ['eid', 'doi', 'pii', 'pubmed_id', 'title', 'subtype', 'subtypeDescription', 'creator', 'afid', 'affilname', 'affiliation_city', 'affiliation_country', 'author_count', 'author_names', 'author_ids', 'author_afids', 'coverDate', 'coverDisplayDate', 'publicationName', 'issn', 'source_id', 'eIssn', 'aggregationType', 'volume', 'issueIdentifier', 'article_number', 'pageRange', 'description', 'authkeywords', 'citedby_count', 'openaccess', 'freetoread', 'freetoreadLabel', 'fund_acr', 'fund_no', 'fund_sponsor']df_results_dia_A.head()| eid | doi | pii | pubmed_id | title | subtype | subtypeDescription | creator | afid | affilname | ... | pageRange | description | authkeywords | citedby_count | openaccess | freetoread | freetoreadLabel | fund_acr | fund_no | fund_sponsor | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2-s2.0-85141971759 | 10.1016/j.neuropharm.2022.109327 | S0028390822003860 | 36368623 | The common genes involved in the pathogenesis ... | ar | Article | Yuan X. | None | Tianjin Medical University | ... | None | None | None | 0 | 0 | None | None | None | None | None |
| 0 | 2-s2.0-85143965932 | 10.1016/j.ebiom.2022.104403 | S2352396422005850 | None | Can the level of copper in the hippocampus wit... | no | Note | Hureau C. | None | Université Fédérale Toulouse Midi-Pyrénées | ... | None | None | None | 0 | 1 | repositoryvor | Green | None | None | None |
| 0 | 2-s2.0-85137505423 | 10.1002/advs.202201882 | None | 36073820 | Hyperglycemic Neurovasculature-On-A-Chip to St... | ar | Article | Jang M. | None | Korea Institute of Science and Technology | ... | None | None | None | 1 | 1 | repositoryvor | Green | None | None | None |
| 0 | 2-s2.0-85143738978 | 10.3390/ijms232315287 | None | 36499613 | Type 2 Diabetes Mellitus and Alzheimer’s Disea... | re | Review | Hamzé R. | None | Université Paris Cité | ... | None | None | None | 0 | 1 | repositoryvor | Green | None | None | None |
| 0 | 2-s2.0-85141352834 | 10.1186/s12987-022-00380-6 | None | 36345028 | Accelerated amyloid angiopathy and related vas... | ar | Article | Vargas-Soria M. | None | Universidad de Cádiz;Instituto de Investigació... | ... | None | None | None | 0 | 1 | repositoryvor | Green | None | None | None |
5 rows × 36 columns
Next, we’ll extract the publication count for three countries - India, China, and United States. Using this data we’ll create a new dataframe having total number of publications based on the two search strategies. In this dataframe, the indicies would be the country name and the columns would be the search keywords.
s1 = (
df_results[df_results["affiliation_country"]
.isin(["India", "United States", "China"])]["affiliation_country"]
.value_counts()
)
s2 = (
df_results_DT[df_results_DT["affiliation_country"]
.isin(["India", "United States", "China"])]["affiliation_country"]
.value_counts()
)
df_final = pd.DataFrame({"Diabetes AND Alzheimer's":s1, "Diabetes AND Parkinson's":s2})
df_final| Diabetes AND Alzheimer's | Diabetes AND Parkinson's | |
|---|---|---|
| United States | 129 | 22 |
| China | 61 | 16 |
| India | 30 | 4 |
Now that we have our dataframe with country-wise number of publications for both the searches, let’s plot this data.
df_final.plot(kind="barh", color=["violet","purple"], \
width=0.5, align="center", figsize=(5,2.25))
plt.xlabel("Total Number of Publications")
ax=plt.gca()
## Replace spaces in the y labels with a new line. ##
labels_edit = []
for l in ax.yaxis.get_ticklabels():
l.set_text(l.get_text().replace(" ", "\n"))
labels_edit.append(l)
ax.yaxis.set_ticklabels(labels_edit)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.yaxis.set_ticks_position('none')
plt.legend(frameon=False, labelcolor=["violet","purple"], \
labelspacing=-2.25)
plt.tight_layout()
plt.savefig("Scopus_search.png", dpi=300)
plt.show()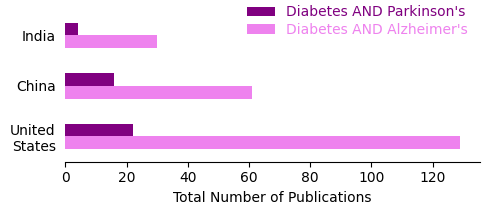
Our search results show that the US is way ahead than the other two countries in terms of publications mentioning Diabetes and Alzheimer’s in the title. These data provides cue to the differences in the research themes internationally in the context of these two disease. Similar differences, albeit less pronounced, can be seen in the publication count with the other search strategy.
Literature mining can provide interesting insight into the publication trends in different contexts. In addition, we can design search strategies to answer specific scientific questions as well. It is no surprise that such text-mining approaches are gaining popularity across disciplines.